Note
Click here to download the full example code
Extract timeseries from ROIs using fmriprep data¶
The first step in most deconvolution analyes is the extraction of the signal from different regions-of-interest.
To do this, nideconv contains an easy-to-use module (nideconv.utils.roi) that leverages the functionality of nilearn. It can extract a time series for every ROI in an atlas. Standard atlases included in nilearn can be found in the nilearn manual.
Using nilearn, the module can also temporally filter the voxelwise signals as well as, clean them from any confounds.
This module is especially useful for preprocessed in the BIDS format, as for example the output of fmriprep.
Extracting a time series from a single functional run¶
Here we extract some signals from a single functional run from a import Stroop dataset we got from a open data repository on Openneuro. The data has been preprocessed using the fmriprep software.
- The raw data was put into /data/openfmri/stroop/sourcedata
- and the data preprocessed with fmriprep in /data/openfmri/stroop/derivatives
# Libraries
from nilearn import datasets
from nideconv.utils import roi
import pandas as pd
from nilearn import plotting
# Locate the data of the first subject
func = '/data/openfmri/ds000164/derivatives/fmriprep/sub-001/func/sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz'
# ... and confounds extracted by fmriprep
confounds_fn = '/data/openfmri/ds000164/derivatives/fmriprep/sub-001/func/sub-001_task-stroop_bold_confounds.tsv'
# We need to load the confounds and fill nas
confounds = pd.read_table(confounds_fn).fillna(method='bfill')
# We only want to include a subset of confounds
confounds_to_include = ['FramewiseDisplacement', 'aCompCor00',
'aCompCor01', 'aCompCor02', 'aCompCor03',
'aCompCor04', 'aCompCor05', 'X', 'Y', 'Z',
'RotX', 'RotY', 'RotZ']
confounds = confounds[confounds_to_include]
# Use the cortical Harvard-Oxford atlas
atlas_harvard_oxford = datasets.fetch_atlas_harvard_oxford('cort-prob-2mm')
plotting.plot_prob_atlas(atlas_harvard_oxford.maps)
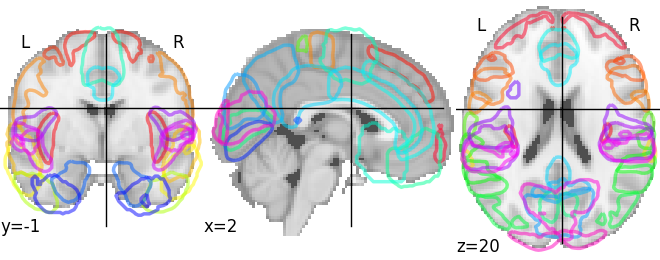
ts = roi.extract_timecourse_from_nii(atlas_harvard_oxford,
func,
confounds=confounds.values,
t_r=1.5,
high_pass=1./128,
)
Now we have a dataframe with a time series for every roi in atlas_harvard_oxford
print(ts.head())
Out:
roi Frontal Pole Insular Cortex Superior Frontal Gyrus ... Planum Temporale Supracalcarine Cortex Occipital Pole
time ...
0.0 0.000682 -0.000467 -0.000190 ... -0.000555 -0.000355 -0.000059
1.5 -0.010150 -0.001433 -0.017112 ... -0.023171 -0.000483 -0.023559
3.0 -0.010898 -0.006012 -0.018928 ... -0.015287 0.002714 -0.016452
4.5 -0.009095 -0.008516 -0.017839 ... -0.026472 0.002852 -0.021837
6.0 -0.006933 -0.006477 -0.014109 ... -0.023391 -0.006571 -0.008051
[5 rows x 48 columns]
An easy way to save these time series is to use the to_pickle functionality of DataFrame
ts.to_pickle('/data/openfmri/stroop_task/derivatives/timeseries/sub-001_task-stroop_harvard_oxford.pkl')
Extract time series for all subjects for complete fmriprep’d dataset¶
nideconv also contains a method to convert an entire fmriprep’d data set to a set of timeseries. This method only needs:
- An atlas in the right format (as supplied with nilearn)
- A BIDS folder containing preprocessed data (e.g., output of fmriprep)
- A BIDS folder containing the raw data.
from nideconv.utils import roi
from nilearn import datasets
# Here we use a subcortical atlas
atlas_pauli = datasets.fetch_atlas_pauli_2017()
plotting.plot_prob_atlas(atlas_pauli)
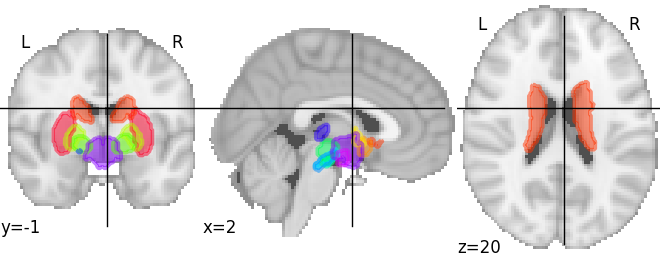
ts = roi.get_fmriprep_timeseries(fmriprep_folder='/data/openfmri/stroop_task/derivatives/fmriprep/',
sourcedata_folder='/data/openfmri/stroop_task/sourcedata/',
atlas=atlas_pauli)
Out:
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-001/func/sub-001_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-002/func/sub-002_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-003/func/sub-003_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-004/func/sub-004_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-005/func/sub-005_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-006/func/sub-006_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-007/func/sub-007_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-008/func/sub-008_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-009/func/sub-009_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-010/func/sub-010_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-011/func/sub-011_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-012/func/sub-012_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-013/func/sub-013_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-014/func/sub-014_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-015/func/sub-015_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-016/func/sub-016_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-017/func/sub-017_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-018/func/sub-018_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-019/func/sub-019_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-020/func/sub-020_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-021/func/sub-021_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-022/func/sub-022_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-023/func/sub-023_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-024/func/sub-024_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-025/func/sub-025_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-026/func/sub-026_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-027/func/sub-027_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
Extracting signal from /data/openfmri/stroop_task/derivatives/fmriprep/sub-028/func/sub-028_task-stroop_bold_space-MNI152NLin2009cAsym_preproc.nii.gz...
We now have a very large dataframe containing time series for every subject and run and for every ROI. The data are indexed with subject and, if applicable session, task, and run.
print(ts)
Out:
roi Pu Ca NAC EXA GPe ... VeP HN HTH MN STH
subject task time ...
001 stroop 0.0 -0.024824 0.005317 0.098381 0.125982 0.091782 ... 0.202659 0.158002 0.023573 0.211872 0.033134
1.5 -0.306827 -0.300921 -0.691543 -1.249696 -0.276813 ... -2.935104 3.526281 0.326159 -0.957822 0.337725
3.0 0.126746 -0.263373 0.828887 -1.982217 -0.148301 ... -1.997062 1.060147 0.368259 0.684619 -0.530833
4.5 -0.058993 -0.064497 0.071342 1.246516 0.630554 ... 1.453879 3.973592 -0.438184 -0.448751 -0.673395
6.0 -0.400270 -0.131084 -0.625979 -0.240803 -0.262864 ... 3.694245 -0.485297 -0.227601 0.494319 -1.236625
7.5 -0.594582 -0.513957 -0.995756 1.024497 -0.293712 ... 0.847354 -4.501187 -0.225672 3.020581 -1.995670
9.0 -0.449662 -0.591477 -1.091159 0.880994 0.410390 ... 0.563932 -2.134154 -0.126763 -0.774114 -1.798392
10.5 -0.208486 -0.381288 -0.112447 0.431814 0.024794 ... -1.370045 -1.974902 -0.281998 1.564937 -0.466028
12.0 -0.343081 -0.225314 -0.189731 -0.972417 -0.743845 ... -2.359034 0.857672 -0.424203 -1.777042 -2.303664
13.5 -0.459105 -0.598026 -0.533962 -1.010868 -0.251734 ... 1.629834 -6.938855 0.692716 0.741312 -1.470699
15.0 -0.491484 -0.327372 0.607643 0.843527 -0.615236 ... 3.607578 1.481565 0.065486 -0.950975 -2.257750
16.5 -0.146404 -0.651250 -0.097022 0.017546 -0.249387 ... 0.728848 -1.740604 -0.143918 -0.533464 -0.344056
18.0 -0.373896 -0.179628 0.044084 0.328200 -0.447820 ... 0.546706 -1.253267 -0.435520 0.604706 -0.854877
19.5 -0.099276 -0.452165 0.160602 -0.710290 -0.554776 ... 0.615395 0.335330 0.658555 -0.805163 -0.434580
21.0 0.045956 -0.176265 -0.144740 -0.156893 0.137828 ... -1.213612 -0.427435 -0.013467 -0.668370 -0.341432
22.5 -0.081434 -0.363184 -0.214064 -0.216374 -0.051227 ... -2.587318 -3.152864 -0.183157 0.876120 -0.876575
24.0 -0.307041 -0.105083 0.264052 -0.353192 -0.095140 ... -1.014375 -0.148635 -0.235113 -2.459859 -0.780580
25.5 -0.383516 -0.345184 0.509282 -1.531761 0.028877 ... 1.643677 -5.526221 -0.104641 0.168232 -0.013299
27.0 -0.267569 -0.117790 0.755515 0.803180 0.629724 ... 1.262726 0.924462 0.100649 1.496751 -0.278082
28.5 -0.361883 -0.334188 -0.116929 1.694318 -0.393364 ... -0.108977 0.734137 -0.267002 -0.631139 -0.790549
30.0 -0.071437 -0.105663 -0.340938 -1.097817 -0.318331 ... 1.783942 -4.189012 0.047490 -0.593999 0.282345
31.5 0.144421 0.009748 -0.505244 -0.636580 -0.118336 ... 2.621462 1.910466 -0.506244 -0.109885 -0.616836
33.0 -0.062435 0.101496 0.533229 -0.499491 0.184077 ... -1.031575 -2.678909 -0.272494 2.096710 -0.417437
34.5 0.070220 -0.026479 -0.251159 0.556795 -0.838313 ... -0.098461 -2.043544 0.267077 -0.305758 -0.157330
36.0 0.080010 -0.178705 -0.350321 -0.854944 0.503817 ... 1.129987 1.555083 -0.257059 -0.804126 1.013317
37.5 -0.072593 0.045305 -0.010711 0.140783 0.209213 ... 3.374004 0.396039 0.258189 -2.795393 -0.161647
39.0 0.156933 -0.269197 -0.585724 -0.596147 -0.137166 ... -1.715686 -4.348575 -0.047123 0.929340 0.358612
40.5 0.171676 -0.118252 0.627424 -1.188610 -0.020654 ... -0.497419 0.466622 -0.168192 -1.940330 -0.409833
42.0 -0.090199 0.020260 0.224642 -0.204530 -0.367158 ... -2.483469 -3.125973 0.709150 0.275251 -0.056099
43.5 -0.006724 0.092758 0.290414 -0.494094 -0.220415 ... -1.992957 2.744478 -0.153984 -3.685024 -1.789079
... ... ... ... ... ... ... ... ... ... ... ...
028 stroop 510.0 -0.000659 -0.100086 -0.662823 -0.322409 0.244794 ... -1.750508 0.651356 -0.188932 0.636989 0.341632
511.5 -0.069122 -0.035227 -0.455429 -0.169374 -0.290590 ... 2.424526 -0.472725 -0.763888 0.687077 -0.905188
513.0 -0.092176 -0.056032 -0.029846 -0.178926 0.465211 ... -0.925284 -1.346232 0.049967 0.748831 1.101769
514.5 -0.001127 -0.167024 -0.535128 0.036451 -0.252076 ... 0.153997 0.369322 -0.571099 0.028998 -0.333085
516.0 -0.039429 -0.040291 -0.106873 -0.646278 -0.834980 ... -1.088412 -0.603679 -0.542698 -1.538116 2.716484
517.5 0.042434 -0.238461 -0.047380 -0.029248 0.296041 ... -1.627384 -3.870078 0.494523 -0.962271 0.790728
519.0 0.048889 -0.088462 -0.111574 -0.615242 -0.192059 ... 0.380134 1.347656 -0.713702 -0.380996 0.982982
520.5 0.056310 -0.005925 0.416474 -1.303348 0.051203 ... 0.231380 -1.882325 -0.163210 0.227902 0.992329
522.0 0.041298 0.022731 -0.033397 -0.009638 0.164090 ... 0.664663 3.253373 -0.069452 1.688823 0.655181
523.5 -0.138554 0.114313 0.185558 0.647344 0.365004 ... -0.366816 2.949501 -0.004114 -0.022473 -0.814738
525.0 -0.077593 0.153181 -0.478577 -0.591277 0.324527 ... -0.650740 1.498044 0.834692 -0.076548 1.264570
526.5 -0.113477 -0.135194 -0.503829 -0.718250 -0.469233 ... 1.008015 -0.894386 -0.502698 1.269312 -0.052719
528.0 -0.033154 0.121374 -0.647306 -0.200863 0.123628 ... -0.752909 -0.989394 0.154674 -0.399817 0.177255
529.5 0.460705 0.050999 -0.372112 0.434588 -0.430714 ... 1.226812 1.699830 -0.474807 0.072520 -0.050545
531.0 0.283997 -0.009623 -0.022878 -0.061585 -0.032400 ... -1.989329 -0.946382 0.485436 2.038023 -1.482713
532.5 0.102716 -0.066149 0.152076 0.228064 -0.855818 ... 3.143413 -2.950538 0.037868 0.464920 0.072040
534.0 -0.318551 -0.158901 -0.327959 0.401055 0.247583 ... -1.053805 -3.719073 -0.159115 0.454866 -0.161271
535.5 -0.662931 -0.265144 0.131707 -0.945303 -0.440436 ... -1.041556 -0.768201 0.373429 -2.610995 -2.657022
537.0 -0.351289 -0.295574 0.455739 -0.098979 -0.371119 ... 2.282096 -0.211732 -0.817694 -2.343147 -1.606882
538.5 0.293919 0.030136 -0.139833 1.120084 0.207005 ... 0.165288 1.321914 0.165129 -0.099461 -1.265270
540.0 0.553214 0.238073 0.364720 1.413141 0.246156 ... -0.215140 -0.939121 0.541443 1.507328 -0.194477
541.5 -0.037640 -0.018405 0.254527 -0.403528 -0.385405 ... 0.118627 -0.955283 0.135546 -0.639177 -0.116389
543.0 -0.303300 -0.109989 0.391246 -0.653175 -0.032351 ... -2.226433 2.809995 0.310023 0.257117 0.573156
544.5 -0.620516 -0.087524 0.217855 -0.070457 -0.086806 ... -0.331080 0.902673 0.287605 -1.478492 -0.899452
546.0 -0.547904 -0.283596 0.100694 0.351159 -0.177932 ... -1.710300 -1.809199 0.504293 1.410871 -0.961112
547.5 -0.397426 -0.314690 0.310524 -0.682668 -0.580231 ... -1.656477 -2.397482 -0.235764 1.354550 -0.387289
549.0 -0.148036 -0.194763 -0.604797 -0.638916 -0.164227 ... -2.277775 -2.042818 -0.085919 0.934715 -0.902066
550.5 -0.112926 -0.329659 -0.119895 -0.432620 -0.303010 ... -2.143460 -6.103886 -0.434857 1.696892 -2.466397
552.0 0.103200 -0.087416 -0.148366 0.119024 -1.550297 ... -1.571636 -1.190581 -0.096551 0.300252 -2.362502
553.5 0.021301 0.021162 0.137266 0.237211 0.057831 ... -0.315415 -0.099618 0.164036 -0.357988 -0.261514
[10360 rows x 16 columns]
(370, 48)
Now we can save these time series for later use.
ts.to_pickle('/data/openfmri/stroop/derivatives/timeseries/pauli_2017.pkl')
Harvard-Oxford atlas¶
For later use, we also extract data using the Harvard-Oxford cortical atlas
atlas_harvard_oxford = datasets.fetch_atlas_harvard_oxford('cort-prob-2mm')
ts = roi.get_fmriprep_timeseries(fmriprep_folder='/data/openfmri/stroop_task/derivatives/fmriprep/',
sourcedata_folder='/data/openfmri/stroop_task/sourcedata/',
atlas=atlas_harvard_oxford)
ts.to_pickle('/data/openfmri/stroop/derivatives/timeseries/harvard_oxford_2017.pkl')
Total running time of the script: ( 0 minutes 0.000 seconds)