Note
Click here to download the full example code
Deconvolution on group of subjects¶
The GroupResponseFitter-object of Nideconv offers an easy way to fit the data of many subjects together well-specified model nideconv.simulate can simulate data from multiple subjects.
from nideconv import simulate
data, onsets, pars = simulate.simulate_fmri_experiment(n_subjects=8,
n_rois=2,
n_runs=2)
Now we have an indexed Dataframe, data, that contains time series for every subject, run, and ROI:
print(data.head())
Out:
area 1 area 2
subject run t
1 1 0.0 0.108141 0.206549
1.0 -0.199233 0.100078
2.0 0.152216 1.322024
3.0 -0.209591 0.410515
4.0 -1.206617 -0.040518
print(data.tail())
Out:
area 1 area 2
subject run t
8 2 295.0 2.797666 2.103899
296.0 -0.308147 0.574413
297.0 0.122700 1.787650
298.0 1.305447 2.825086
299.0 1.271533 1.641312
We also have onsets for every subject and run:
print(onsets.head())
Out:
onset
subject run trial_type
1 1 Condition A 219.820716
Condition A 103.107395
Condition A 266.262272
Condition A 7.918901
Condition A 89.896628
print(onsets.tail())
Out:
onset
subject run trial_type
8 2 Condition B 182.382405
Condition B 237.654933
Condition B 258.877295
Condition B 63.358865
Condition B 37.917893
We can now use GroupResponseFitter to fit all the subjects with one object:
from nideconv import GroupResponseFitter
# `concatenate_runs` means that we concatenate all the runs of a single subject
# together, so that we have to fit only a single GLM per subject.
# A potential advantage is that the GLM has less degrees-of-freedom
# compared to the amount of data, so that the esitmate are potentially more
# stable.
# A potential downside is that different runs might have different intercepts
# and/or correlation structure.
# Therefore, by default, the `GroupResponseFitter` does _not_ concatenate
# runs.
g_model = GroupResponseFitter(data,
onsets,
input_sample_rate=1.0,
concatenate_runs=False)
We use the add_event-method to add the events we are interested in. The GroupResponseFitter then automatically collects the right onset times from the onsets-object.
We choose here to use the Fourier-basis set, with 9 regressors.
g_model.add_event('Condition A',
basis_set='fourier',
n_regressors=9,
interval=[0, 20])
g_model.add_event('Condition B',
basis_set='fourier',
n_regressors=9,
interval=[0, 20])
We can fit all the subjects at once using the fit-method
g_model.fit()
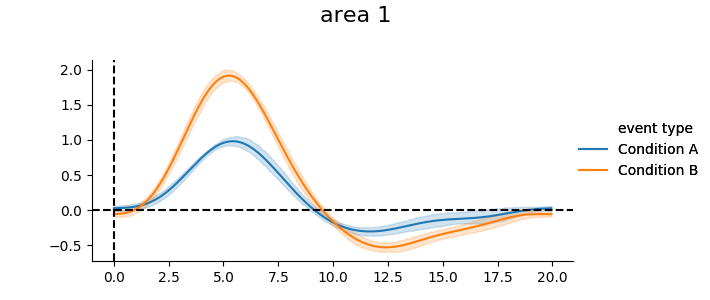
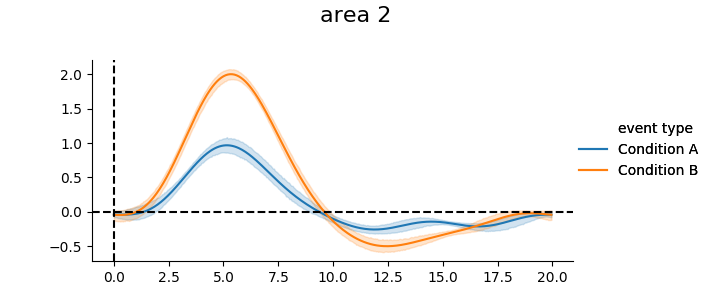
We can plot the mean timecourse across subjects
print(g_model.get_subjectwise_timecourses().head())
g_model.plot_groupwise_timecourses()
Out:
area 1 area 2
subject event type covariate time
1 Condition A intercept 0.00 0.056636 0.047815
0.05 0.056482 0.047954
0.10 0.056218 0.047809
0.15 0.055854 0.047394
0.20 0.055398 0.046727
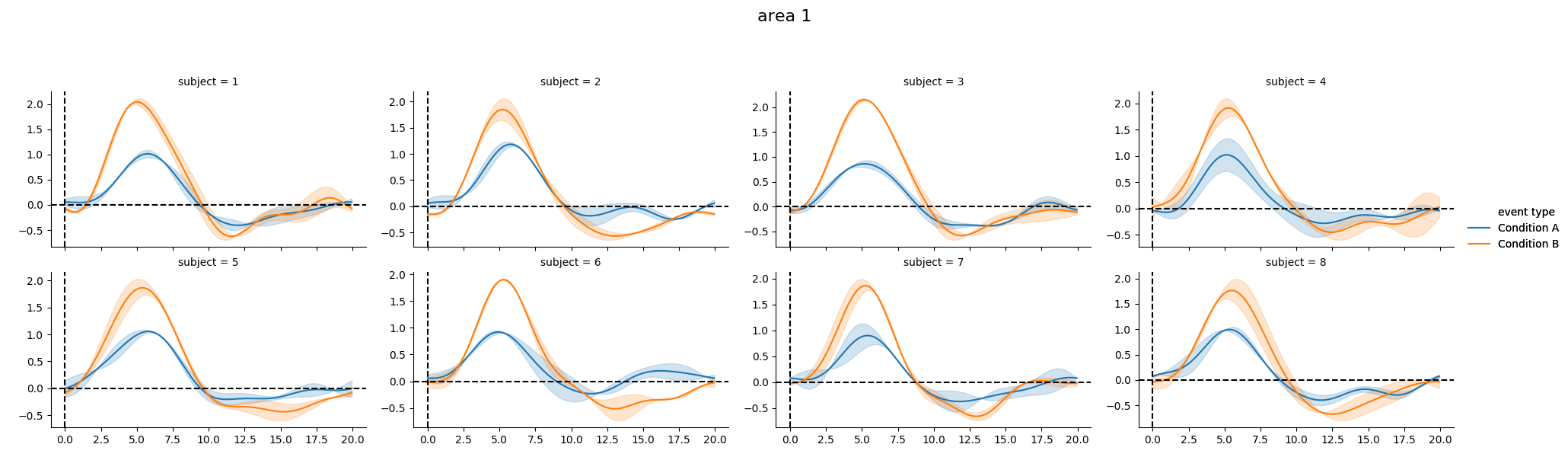
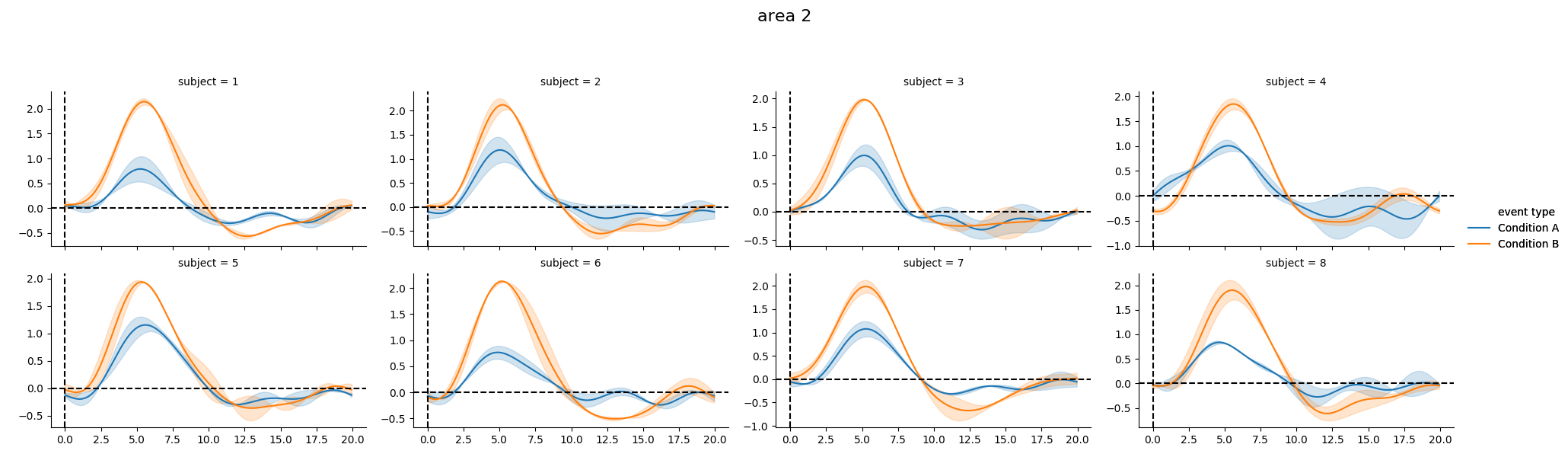
As well as individual time courses
print(g_model.get_conditionwise_timecourses())
g_model.plot_subject_timecourses()
Out:
area 1 area 2
event type covariate time
Condition A intercept 0.00 0.027545 -0.039614
0.05 0.028095 -0.039715
0.10 0.028649 -0.039852
0.15 0.029217 -0.040009
0.20 0.029811 -0.040169
0.25 0.030440 -0.040316
0.30 0.031117 -0.040429
0.35 0.031852 -0.040491
0.40 0.032659 -0.040482
0.45 0.033549 -0.040383
0.50 0.034535 -0.040174
0.55 0.035629 -0.039835
0.60 0.036845 -0.039346
0.65 0.038195 -0.038685
0.70 0.039693 -0.037834
0.75 0.041352 -0.036772
0.80 0.043184 -0.035478
0.85 0.045204 -0.033932
0.90 0.047423 -0.032116
0.95 0.049855 -0.030010
1.00 0.052513 -0.027594
1.05 0.055408 -0.024851
1.10 0.058554 -0.021763
1.15 0.061962 -0.018312
1.20 0.065644 -0.014483
1.25 0.069611 -0.010259
1.30 0.073873 -0.005627
1.35 0.078442 -0.000571
1.40 0.083326 0.004920
1.45 0.088535 0.010859
... ... ...
Condition B intercept 18.50 -0.074890 -0.028460
18.55 -0.072558 -0.026545
18.60 -0.070388 -0.024846
18.65 -0.068382 -0.023366
18.70 -0.066540 -0.022104
18.75 -0.064863 -0.021059
18.80 -0.063349 -0.020230
18.85 -0.061996 -0.019614
18.90 -0.060803 -0.019208
18.95 -0.059764 -0.019008
19.00 -0.058876 -0.019006
19.05 -0.058132 -0.019198
19.10 -0.057526 -0.019574
19.15 -0.057051 -0.020126
19.20 -0.056697 -0.020843
19.25 -0.056454 -0.021716
19.30 -0.056312 -0.022730
19.35 -0.056259 -0.023874
19.40 -0.056281 -0.025132
19.45 -0.056366 -0.026488
19.50 -0.056498 -0.027927
19.55 -0.056661 -0.029431
19.60 -0.056838 -0.030981
19.65 -0.057011 -0.032557
19.70 -0.057162 -0.034138
19.75 -0.057272 -0.035704
19.80 -0.057319 -0.037232
19.85 -0.057283 -0.038699
19.90 -0.057142 -0.040080
19.95 -0.056873 -0.041351
[800 rows x 2 columns]
Total running time of the script: ( 3 minutes 28.376 seconds)